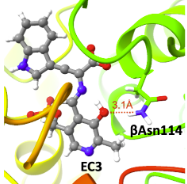
|
Data from simulations of mutations of tryptophan synthase..
References:
A Molecular Dynamics Simulation Study of the Effects of βGln114 Mutation on the Dynamic Behavior of the Catalytic Site of the Tryptophan Synthase. Ry, Anupom; Karttunen, Mikko. JCIM (2024).
https://doi.org/10.1021/acs.jcim.3c01966
|
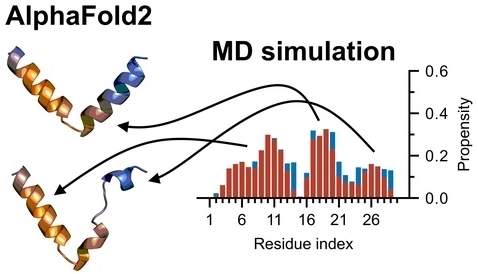
|
Alphafold disordered protein data. Data from the paper below.
References:
AlphaFold2: A Role for Disordered Protein/Region Prediction?. Wilson, Carter J.; Choy, Wing-Yiu; Karttunen, Mikko. Int. J. Mol. Sci. 23, 4591, (2022).
https://doi.org/10.3390/ijms23094591
|
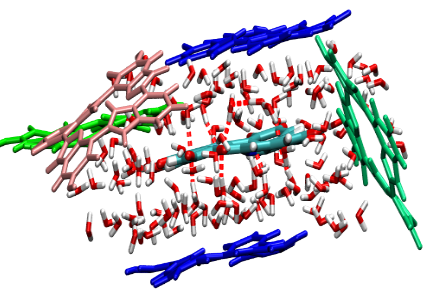
|
Parameters for eumelanin; DHI and DHICA forms
References:
Free Energy and Stacking of Eumelanin Nanoaggregates. Soltani, Sepideh; Sowlati-Hashjin, Shahin; Tetsassi Feugmo, Conrard Giresse; Karttunen, Mikko. J. Phys. Chem. B 126, 1805–1818, (2022).
https://doi.org/10.1021/acs.jpcb.1c07884 Structural Investigation of DHICA Eumelanin Using Density Functional Theory and Classical Molecular Dynamics Simulations. Soltani, Sepideh; Sowlati-Hashjin, Shahin; Tetsassi Feugmo, Conrard Giresse; Karttunen, Mikko. Molecules 27, 8417 (2022).
https://doi.org/10.3390/molecules27238417
|
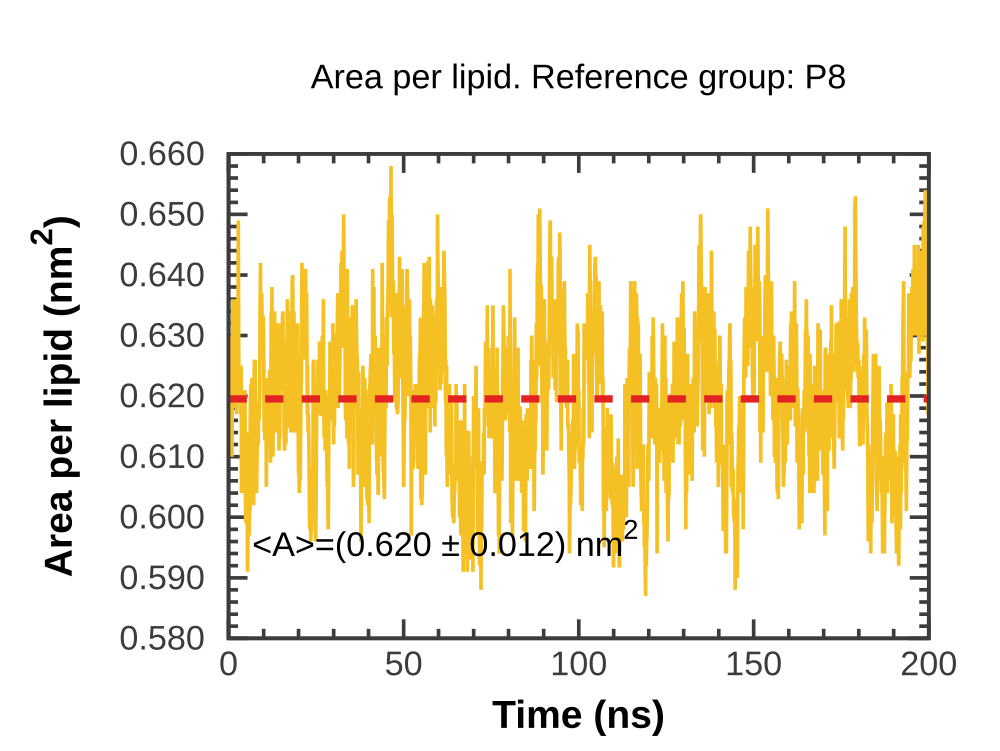
|
DMPC: 200 ns simulation of a DMPC bilayer using Gromos 53A6 + Berger lipids. 128 lipids. |
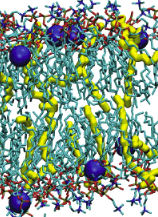
|
DMPC and DMTAP lipids: Configurations and parameters and lipid mixtures. |

|
DPPC: bilayer after 100 ns. |
|
NaCl force field study: All the radial distribution functions and final configurations (PDB). |
|
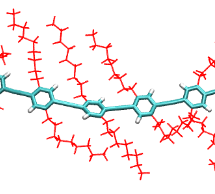
Optimized structures of DPE (diphenylethyne) and friends.
Optimized structures of DPE (diphenylethyne), Me-DPE and n-PPE (n=1,2,…,10) (MD and DFT) and input files for the VOTCA package for calculating excitation energies.
References:
Getting Excited: Challenges in Quantum-Classical Studies of Excitons in Polymeric Systems. Bagheri, Behnaz; Baumeier, Björn; Karttunen, Mikko. Phys. Chem. Chem. Phys. 18, 30297–30304, (2016).
https://doi.org/10.1039/c6cp02944b. Solvent Effects on Optical Excitations of Poly Para Phenylene Ethynylene Studied by QM/MM Simulations Based on Many-Body Green’s Functions Theory. Bagheri, B.; Karttunen, M.; Baumeier, B. Eur. Phys. J. Spec. Top. 225, 1743–1756, (2016).
https://doi.org/10.1140/epjst/e2016-60144-5.
|
|
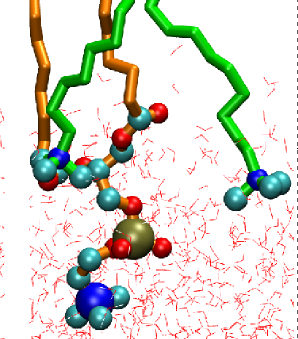
CTAB (cetyltrimethylammonium bromide) parameters.
CTAB is a cationic surfactant. The parameterizaton is based on quantum chemistry calculations and NPA analysis. Download the configurations and parameters here:
- CTAB parameters (ctab.itp)
- Configuration at the end of a 1 microsecond trajectory/ 50% DPPC, 50% CTAB, area per lipid: 0.5 nm2
- README file
References:
Molecular Dynamics Simulations of DPPC/CTAB Monolayers at the Air/Water Interface. Liu, Bin; Hoopes, Matthew I.; Karttunen, Mikko. J. Phys. Chem. B 118, 11723-11737, (2014).
https://doi.org/10.1021/jp5050892
|
|
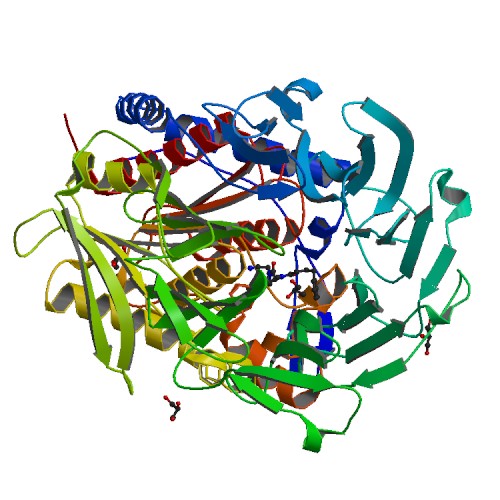
Prolyl oligopeptidase (POP) PDB structures with inhibitors. X-ray diffraction structures
Download the PDB files here:
- 4AMY: Prolyl Oligopeptidase from porcine brain with a covalently bound inhibitor IC-1
- 4AMZ: Prolyl Oligopeptidase from porcine brain with a covalently bound inhibitor IC-2
- 4AN0: Prolyl Oligopeptidase from porcine brain with a covalently bound inhibitor IC-3
- 4AN1: Prolyl Oligopeptidase from porcine brain with a covalently bound inhibitor IC-4
References:
Molecular Dynamics, Crystallography and Mutagenesis Studies on the Substrate Gating Mechanism of Prolyl Oligopeptidase. Kaszuba, Karol; Róg, Tomasz; Danne, Reinis; Canning, Peter; Fülöp, Vilmos; Juhász, Tünde; Szeltner, Zoltán; Pierre, J. F. St; García-Horsman, Arturo; Männistö, Pekka T.; Karttunen, Mikko; Hokkanen, Jyrki; Bunker, Alex. Biochimie 94, 1398–1411, (2012).
https://doi.org/10.1016/j.biochi.2012.03.012.
|








